Publications
Highlighted
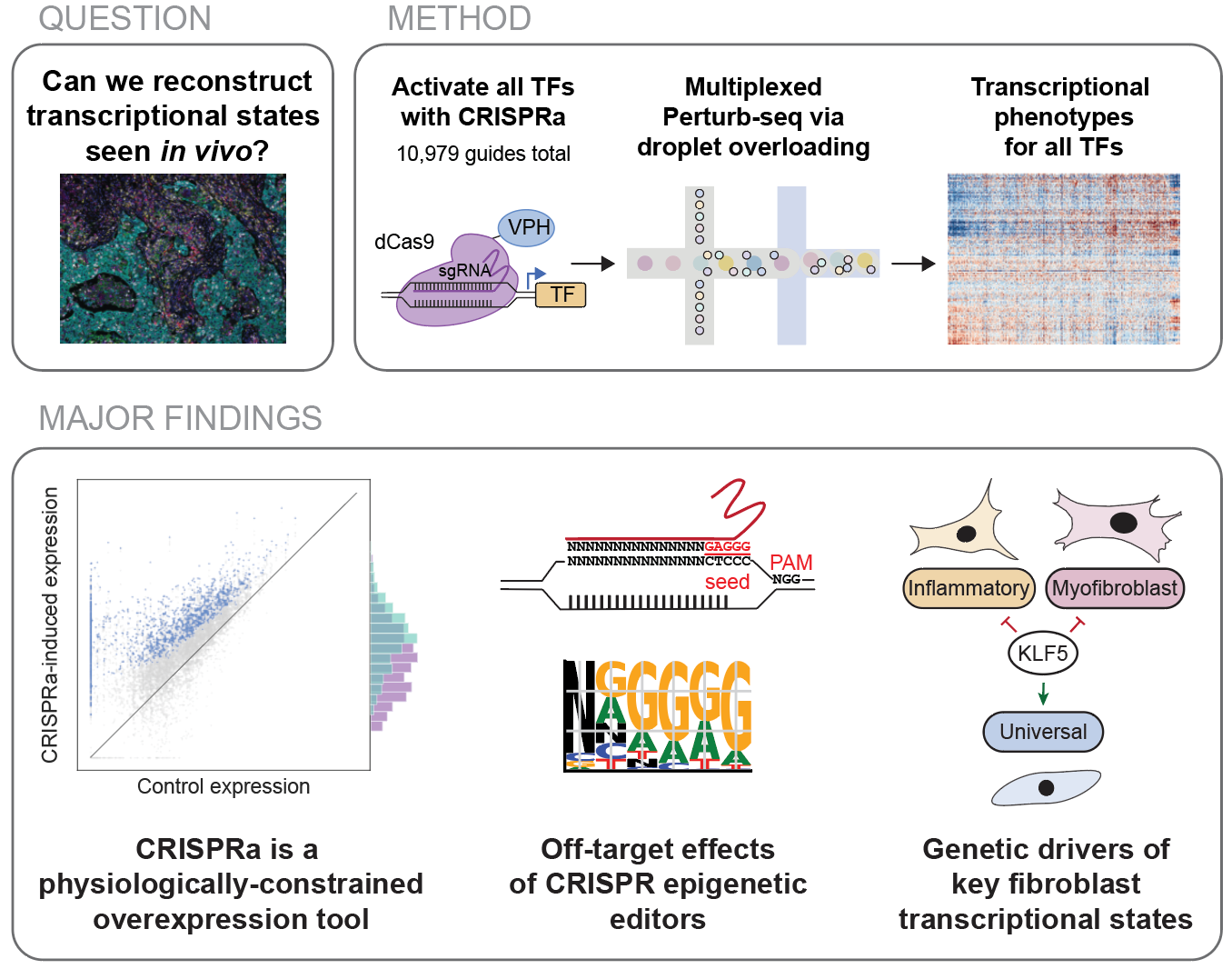
Comprehensive transcription factor perturbations recapitulate fibroblast transcriptional states
bioRxiv
·
03 Aug 2024
·
doi:10.1101/2024.07.31.606073
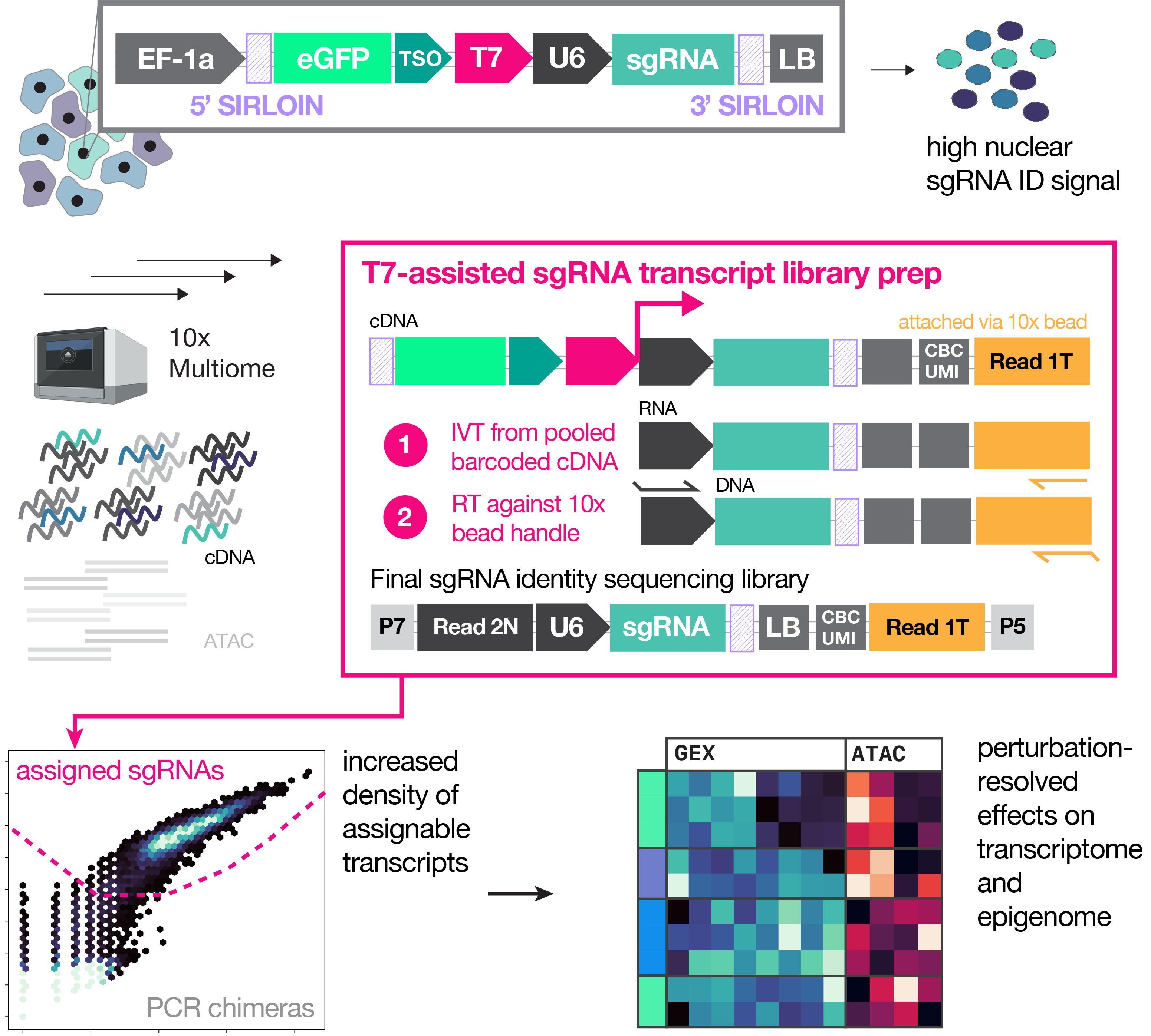
Multiome Perturb-seq unlocks scalable discovery of integrated perturbation effects on the transcriptome and epigenome
Cell Systems
·
01 Jan 2025
·
doi:10.1016/j.cels.2024.12.002
Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq
Cell
·
01 Jul 2022
·
doi:10.1016/j.cell.2022.05.013
All
2025

Multiome Perturb-seq unlocks scalable discovery of integrated perturbation effects on the transcriptome and epigenome
Cell Systems
·
01 Jan 2025
·
doi:10.1016/j.cels.2024.12.002
2024

Comprehensive transcription factor perturbations recapitulate fibroblast transcriptional states
bioRxiv
·
03 Aug 2024
·
doi:10.1101/2024.07.31.606073
Multiome Perturb-seq unlocks scalable discovery of integrated perturbation effects on the transcriptome and epigenome
Cold Spring Harbor Laboratory
·
27 Jul 2024
·
doi:10.1101/2024.07.26.605307
2023
Drivers of heterogeneity in synovial fibroblasts in rheumatoid arthritis
Nature Immunology
·
05 Jun 2023
·
doi:10.1038/s41590-023-01527-9
2022
Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq
Cell
·
01 Jul 2022
·
doi:10.1016/j.cell.2022.05.013
High-content CRISPR screening
Nature Reviews Methods Primers
·
10 Feb 2022
·
doi:10.1038/s43586-021-00093-4
2020
Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing
Nature Biotechnology
·
30 Mar 2020
·
doi:10.1038/s41587-020-0470-y
Titrating gene expression using libraries of systematically attenuated CRISPR guide RNAs
Nature Biotechnology
·
13 Jan 2020
·
doi:10.1038/s41587-019-0387-5
2019
Stochastic antagonism between two proteins governs a bacterial cell fate switch
Science
·
04 Oct 2019
·
doi:10.1126/science.aaw4506
Exploring genetic interaction manifolds constructed from rich single-cell phenotypes
Science
·
23 Aug 2019
·
doi:10.1126/science.aax4438
Molecular recording of mammalian embryogenesis
Nature
·
13 May 2019
·
doi:10.1038/s41586-019-1184-5
2016
Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens
Cell
·
01 Dec 2016
·
doi:10.1016/j.cell.2016.11.038
A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response
Cell
·
01 Dec 2016
·
doi:10.1016/j.cell.2016.11.048
Exploiting Natural Fluctuations to Identify Kinetic Mechanisms in Sparsely Characterized Systems
Cell Systems
·
01 Apr 2016
·
doi:10.1016/j.cels.2016.04.002
Constraints on Fluctuations in Sparsely Characterized Biological Systems
Physical Review Letters
·
01 Feb 2016
·
doi:10.1103/PhysRevLett.116.058101
2013
Memory and modularity in cell-fate decision making
Nature
·
01 Nov 2013
·
doi:10.1038/nature12804
2011
Evidence that metabolism and chromosome copy number control mutually exclusive cell fates inBacillus subtilis
The EMBO Journal
·
15 Feb 2011
·
doi:10.1038/emboj.2011.36
2010
Broadly heterogeneous activation of the master regulator for sporulation in
Bacillus subtilis
Proceedings of the National Academy of Sciences
·
19 Apr 2010
·
doi:10.1073/pnas.1002499107
An epigenetic switch governing daughter cell separation in Bacillus subtilis
Genes & Development
·
29 Mar 2010
·
doi:10.1101/gad.1915010